1. Introduction.
PST-PRNA is a computable method for predicting RNA binding sites on proteins.
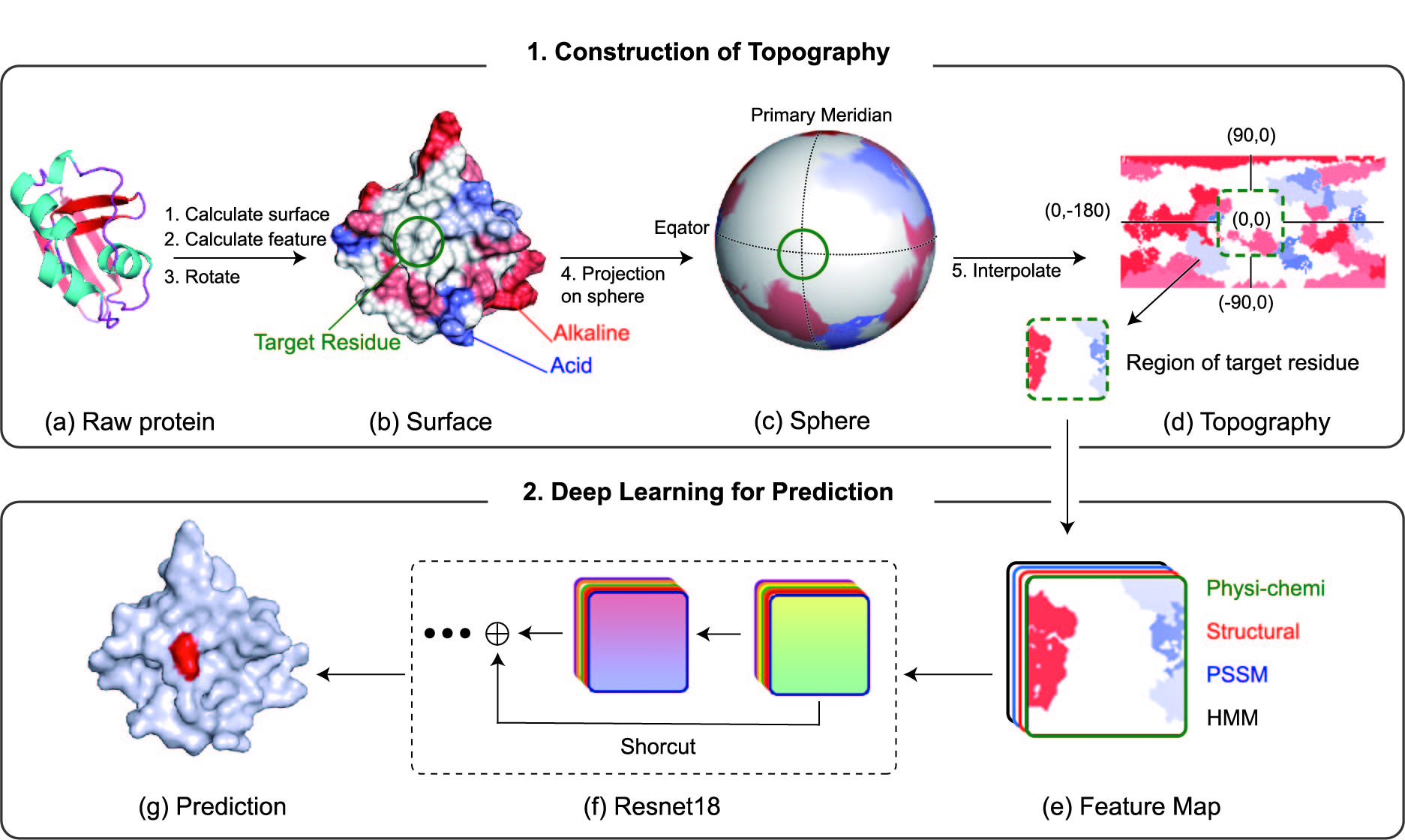
Two steps are performed in PST-PRNA. The first step is generating Topography. The second step is training and predicting by using a deep learning model.
PST-PRNA is a computable method for predicting RNA binding sites on proteins.
Two steps are performed in PST-PRNA. The first step is generating Topography. The second step is training and predicting by using a deep learning model.

Upload the PDB format file of protein structure. The file can only contain one protein chain to be analyzed.
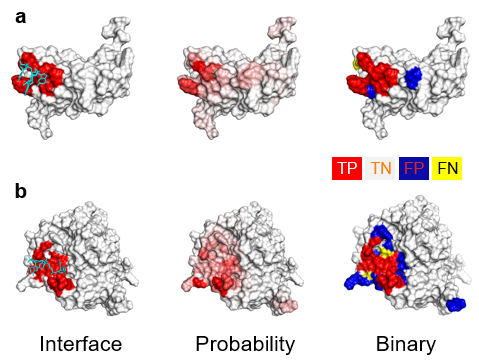
The results yield the predicted binding score for each residue located on the surface of protein.
a. PST-PRNA works on the surface of protein.
Residues hidden inside of the protein are labeled as i (internal) and the RNA binding preference is 0.
Residues exposed to the surface are predicted by deep learning model and given the binding preference score.
b. PST-PRNA relies less on sequence identity.
In related works, sequence-based methods rely too much on sequence similarity. And due to that, at the process of data preprocessing,
those methods generally assess the sequence identity of dataset and remove proteins whose sequence identities are higher than 0.3. We compare PST-PRNA with our previous work PRNA which represents sequence-based methods on different sequence identities. The result shows that PST-PRNA can overcome the shortcoming of sequence similarity and learns the latent structure information.
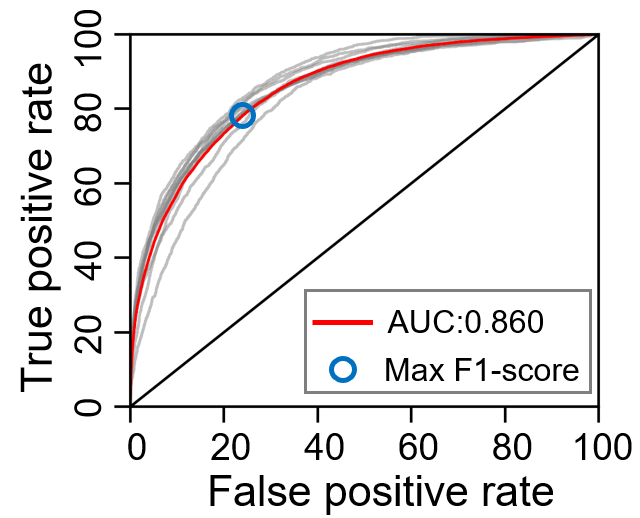
PST-PRNA gives the bindingg preference of one residue on the protein surface. Users could discriminate one site binding or not according the preference As for making a binary classification, we give the ROC of PST-PRNA for choosing the threshold.
The dot nearest to the left-top is considered to be the best performance for the model. And when threshold is 0.129, the sensitivity is 0.783 and specificity is 0.760.